import pyscan
import matplotlib.pyplot as plt
import random
import math
import statistics
import csv
import itertools
import numpy as np
def plot_points(ax, pts, c):
xs = []
ys = []
for pt in pts:
xs.append(pt[0] )
ys.append(pt[1])
ax.scatter(xs, ys, color=c)
def plot_points_traj(ax, pts, c):
xs = []
ys = []
for pt in pts:
xs.append(pt[0])
ys.append(pt[1])
ax.plot(xs, ys, color=c)
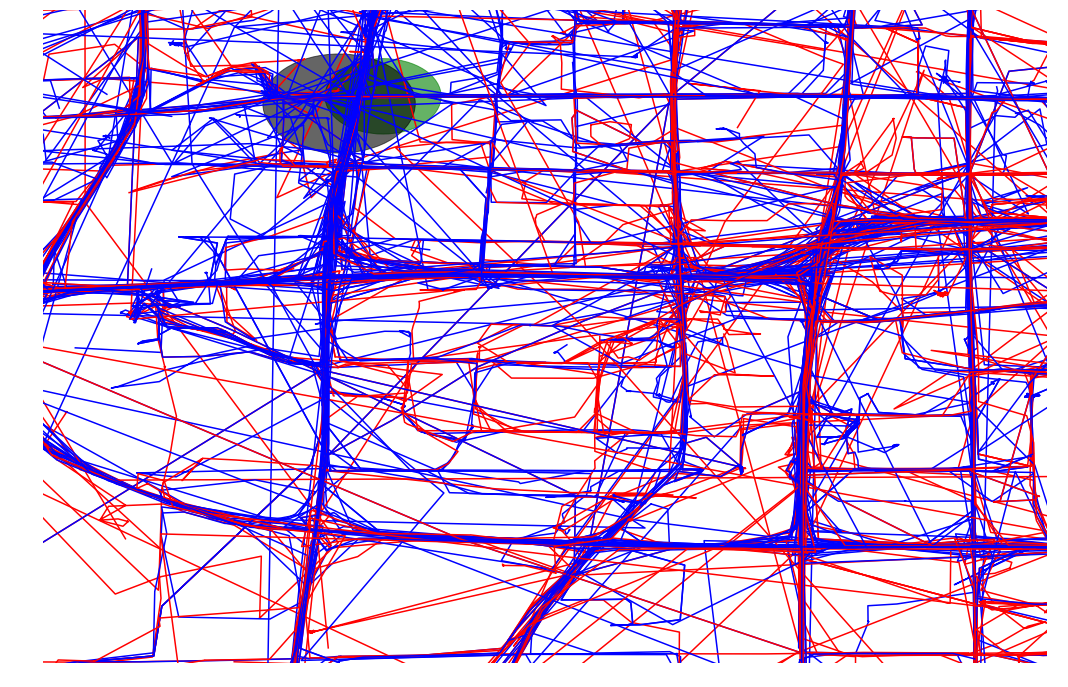
def plot_approx(ax, traj_pts, core_set_pts):
ax.set_xlim([-.01, 1.01])
ax.set_ylim([-.01, 1.01])
plot_points_traj(ax, traj_pts, "g")
plot_points(ax, core_set_pts, "b")
ax.set_axis_off()
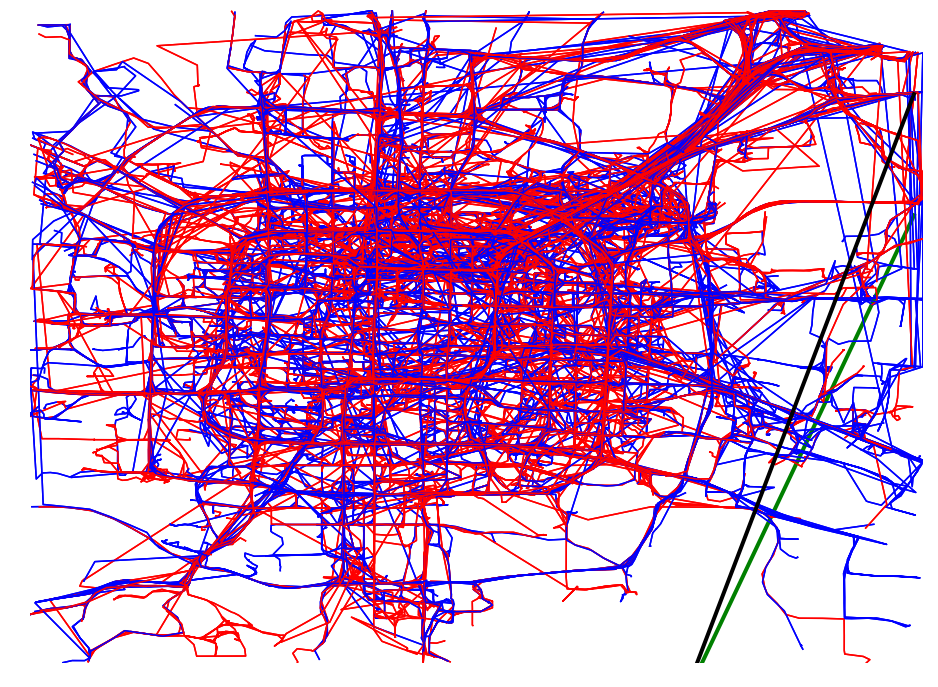
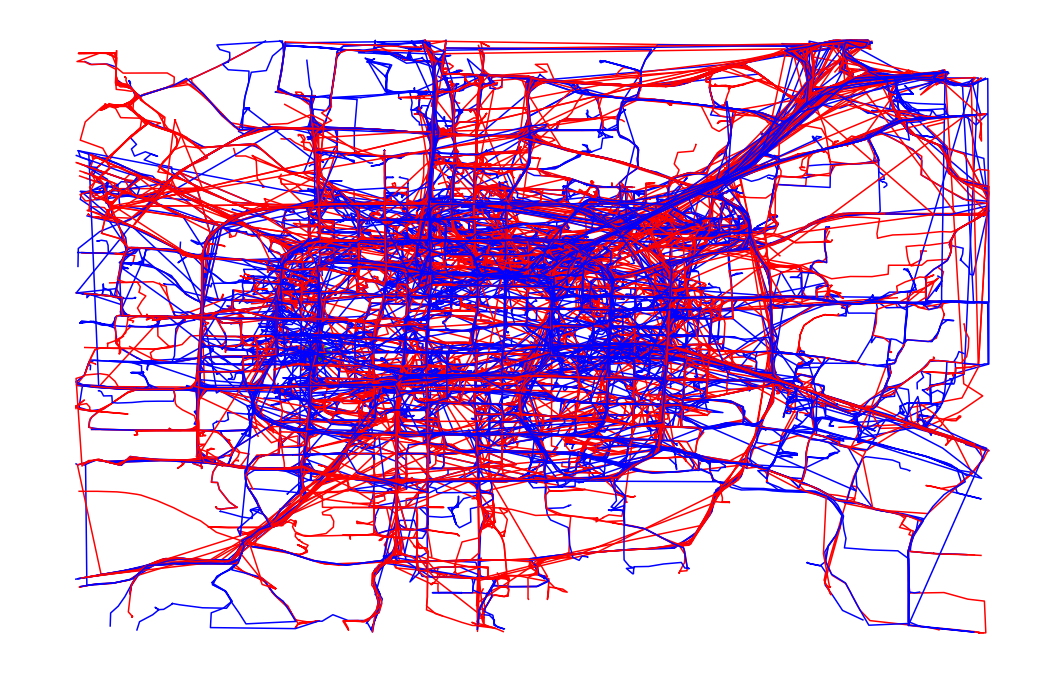
def plot_full_trajectories(red, blue, ax):
plot_set = [(reg, True) for reg in blue] + [(reg, False) for reg in red]
random.shuffle(plot_set)
for traj, is_blue in plot_set:
if is_blue:
plot_points_traj(ax, traj, "b")
else:
plot_points_traj(ax, traj, "r")
def remove_long_trajectories(trajectories, percent=.9):
def toTraj(pts):
return pyscan.Trajectory([pyscan.Point(p[0], p[1], 1.0) for p in pts])
ltraj = sorted(toTraj(traj).get_length() for traj in trajectories)
perc_len_traj = ltraj[int(percent * len(trajectories))]
del ltraj
return [traj for traj in trajectories if toTraj(traj).get_length() <= perc_len_traj]
def normalize(pt, mxx, mnx, mxy, mny):
return (pt[0] - mnx) / (mxx - mnx), (pt[1] - mny) / (mxy - mny)
def read_csv(fname, filter_long=True):
traj_id_set = []
with open(fname) as f:
curr_id = None
mnx = float("inf")
mxx = -float("inf")
mny = float("inf")
mxy = -float("inf")
for row in csv.reader(f, delimiter=" "):
try:
if row[0] != curr_id:
curr_id = row[0]
traj_id_set.append([])
if math.isnan(float(row[1])) or math.isinf(float(row[1])):
continue
if math.isnan(float(row[2])) or math.isinf(float(row[2])):
continue
x, y = (float(row[1]), float(row[2]))
mnx = min(mnx, x)
mny = min(mny, y)
mxx = max(mxx, x)
mxy = max(mxy, y)
traj_id_set[-1].append((float(row[1]), float(row[2])))
except ValueError:
continue
norm_traces = []
while traj_id_set:
norm_trace = []
trace = traj_id_set.pop()
for pt in trace:
norm_trace.append(normalize(pt, mxx, mnx, mxy, mny))
norm_traces.append(norm_trace)
if filter_long:
return remove_long_trajectories(norm_traces)
else:
return norm_traces
 pyscan
1.0
pyscan
1.0