import pyscan
import matplotlib.pyplot as plt
import random
import math
import statistics
def plot_points(ax, pts, c):
xs = []
ys = []
for pt in pts:
xs.append(pt[0] )
ys.append(pt[1])
ax.scatter(xs, ys, color=c)
def plot_points_traj(ax, pts, c):
xs = []
ys = []
for pt in pts:
xs.append(pt[0])
ys.append(pt[1])
ax.plot(xs, ys, color=c)
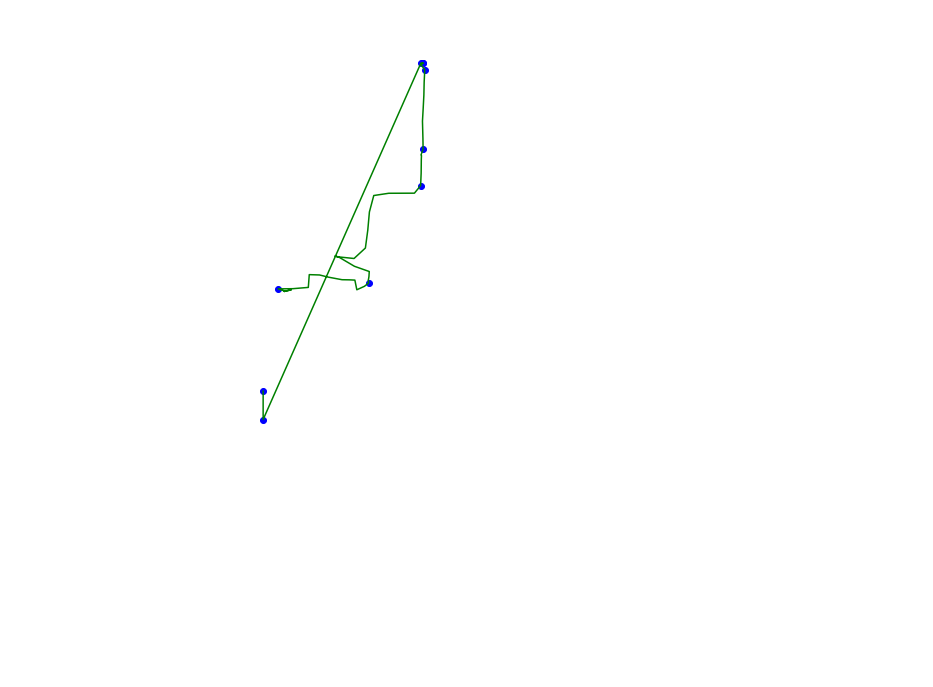
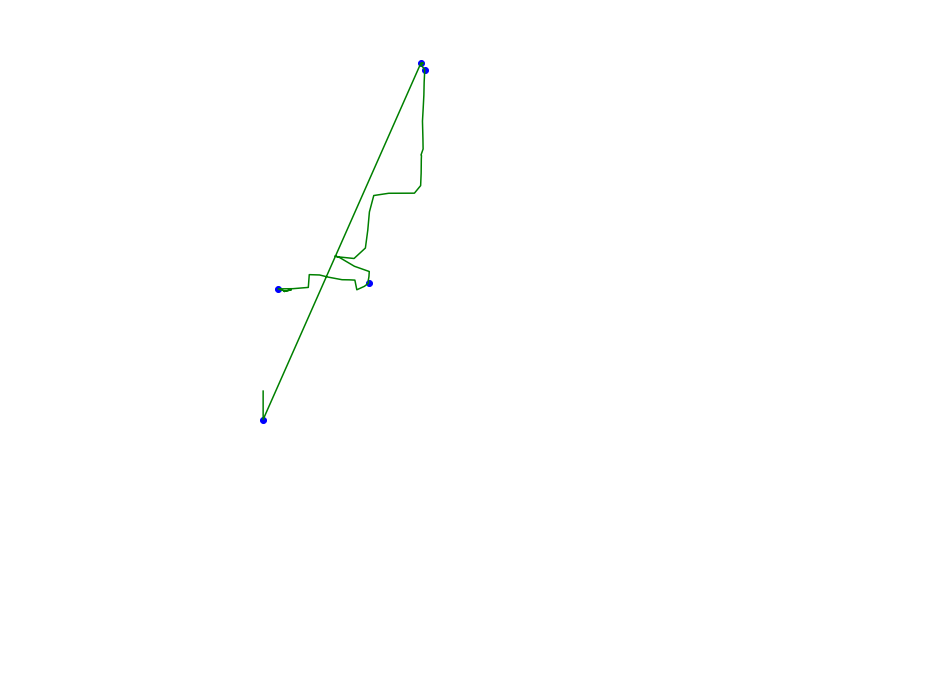
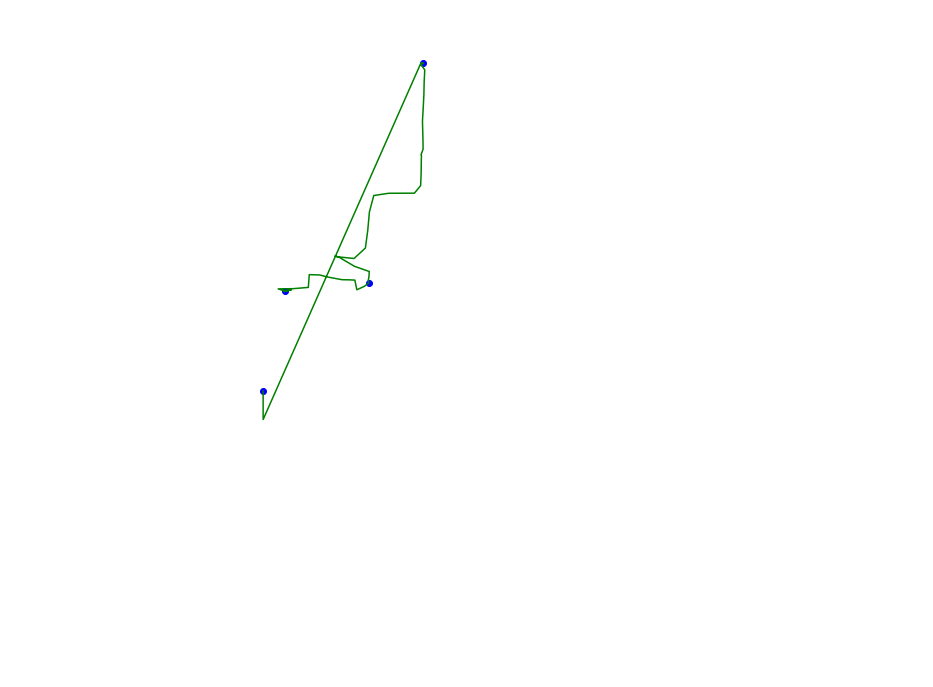
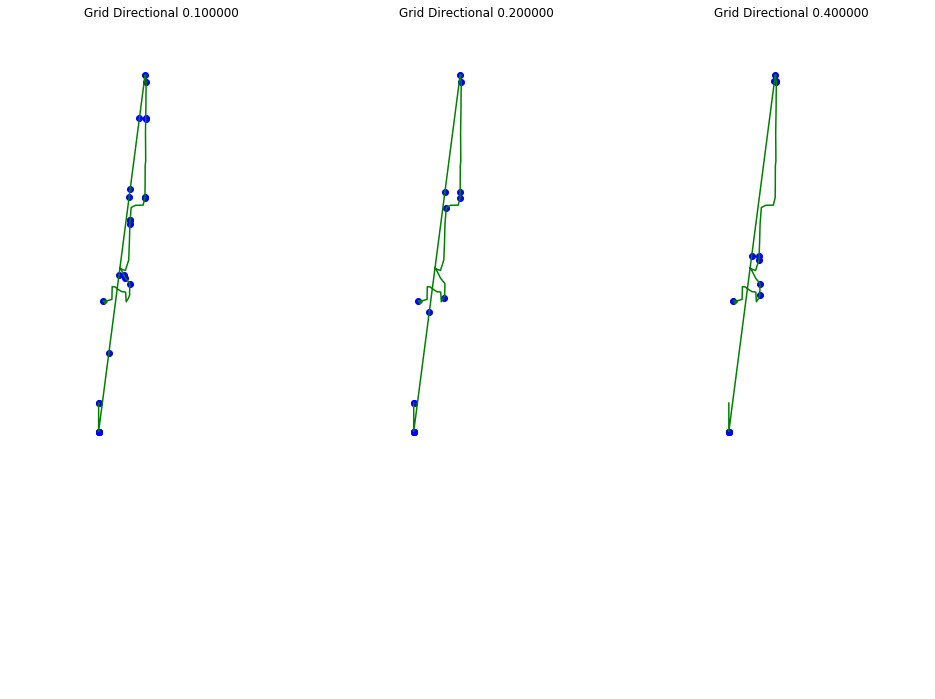
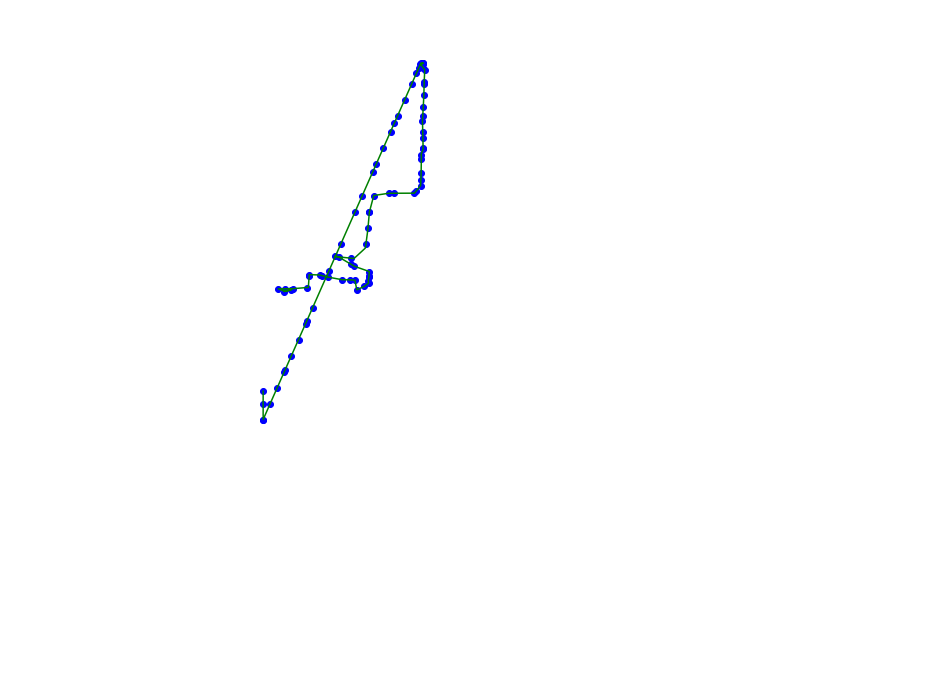
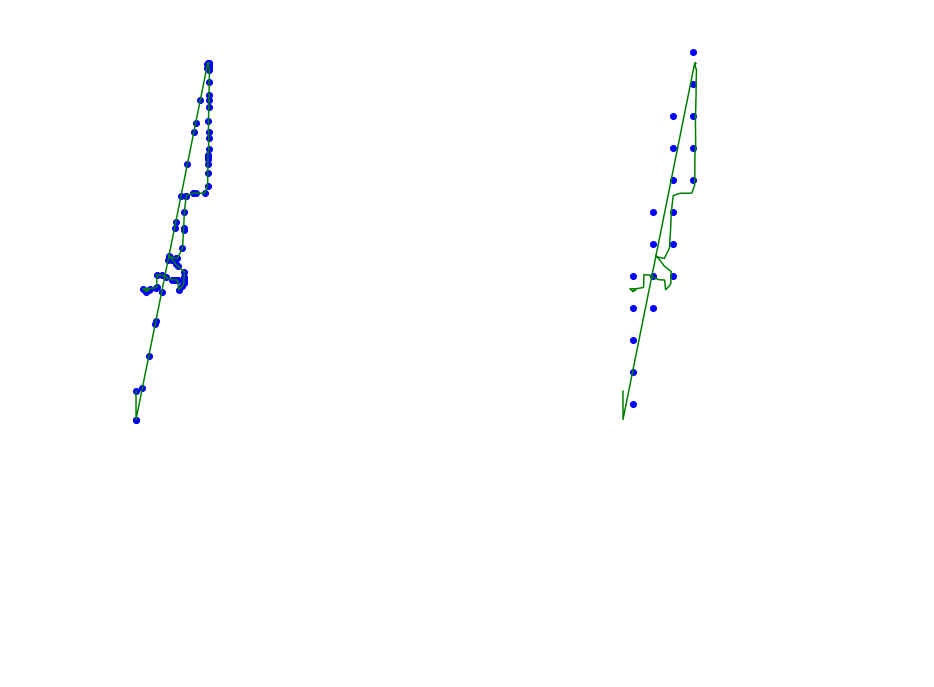
def plot_approx(ax, traj_pts, core_set_pts):
ax.set_xlim([-.01, 1.01])
ax.set_ylim([-.01, 1.01])
plot_points_traj(ax, traj_pts, "g")
plot_points(ax, core_set_pts, "b")
ax.set_axis_off()
pts = [pyscan.Point(x, y, 1.0) for (x, y) in (
(0.28076897884325597, 0.5709315642759948),
(0.2883670662212526, 0.5731789009659035),
(0.2804742254535915, 0.5708019102361805),
(0.28057247658348744, 0.5704993841433011),
(0.2807034780899998, 0.5714501804351904),
(0.2734656448549006, 0.5746699224236597),
(0.2902993384423915, 0.5749940575231799),
(0.3078862906923401, 0.5769820861334908),
(0.30896705312110206, 0.5969488082632861),
(0.32072443833104086, 0.5964301921040598),
(0.3307133032029929, 0.5930375780625253),
(0.34640073360844237, 0.5890183028286218),
(0.35494858190868855, 0.588802212762275),
(0.3610729023383628, 0.5883916416362222),
(0.36329992794916616, 0.5732869459990769),
(0.37201152813256444, 0.5787108066642039),
(0.37682583349709176, 0.5840266222961576),
(0.3765310801074273, 0.5862307409728461),
(0.37640007860089164, 0.5878730254770267),
(0.3764328289775314, 0.5880675065367327),
(0.37649832973078756, 0.5883700326295813),
(0.37649832973078756, 0.5889750848153402),
(0.3765310801074273, 0.5894504829612848),
(0.3765310801074273, 0.5894504829612848),
(0.3765310801074273, 0.5894504829612848),
(0.3766293312372999, 0.5896017460077398),
(0.3771533372633727, 0.5939019383278819),
(0.3775135914062933, 0.6018324437625461),
(0.3605816466889298, 0.610000648270198),
(0.34266719067270024, 0.6244570737083028),
(0.3384096417108854, 0.6261857942390467),
(0.3402764131787448, 0.6249972988741699),
(0.36012314141611323, 0.6221881280117534),
(0.3730922905613496, 0.638567755040308),
(0.37581057182156274, 0.6664649826047369),
(0.3776773432894454, 0.6949456533483143),
(0.38265540053710156, 0.7206603712427262),
(0.3997510971376172, 0.7241826393240749),
(0.42909543459750465, 0.7243339023704993),
(0.43620226632606823, 0.7361540289992835),
(0.43679177310539713, 0.7557750070229177),
(0.4369882753651657, 0.7778810208094789),
(0.4371520272483178, 0.7831752274347917),
(0.43662802122224503, 0.7839963696868667),
(0.4390187987161772, 0.7933962875726462),
(0.4388222964564087, 0.809970395660907),
(0.4382982904303359, 0.8370032629599792),
(0.4391825505993293, 0.8585690515807041),
(0.4399030588851706, 0.8777146314583937),
(0.4402305626514748, 0.8978758346478641),
(0.44091832056067637, 0.9170862415454762),
(0.43639876858583676, 0.925729844199042),
(0.4386912949498963, 0.9279555718823406),
(0.43878954607976894, 0.9269183395639188),
(0.43875679570315246, 0.9269399485705596),
(0.43875679570315246, 0.9269615575772004),
(0.43872404532651277, 0.9269831665838106),
(0.43872404532651277, 0.9270263845970922),
(0.43872404532651277, 0.9269183395639188),
(0.43878954607976894, 0.9269399485705596),
(0.4388550468330484, 0.9274153467165043),
(0.43675902272875744, 0.9279987898956222),
(0.4359402633130202, 0.9259675432720297),
(0.2562716971245123, 0.37042159171941874),
(0.2562716971245123, 0.4148497093588573))]
alpha = 1/20
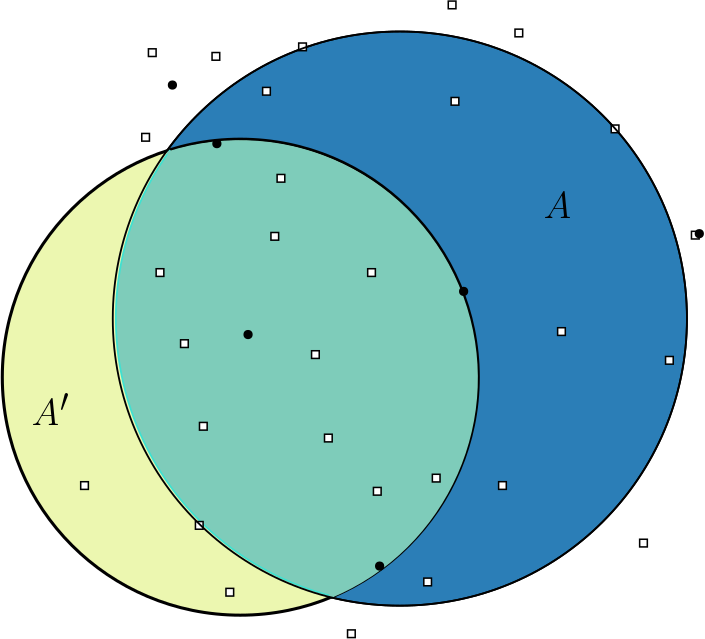 pyscan
1.0
pyscan
1.0