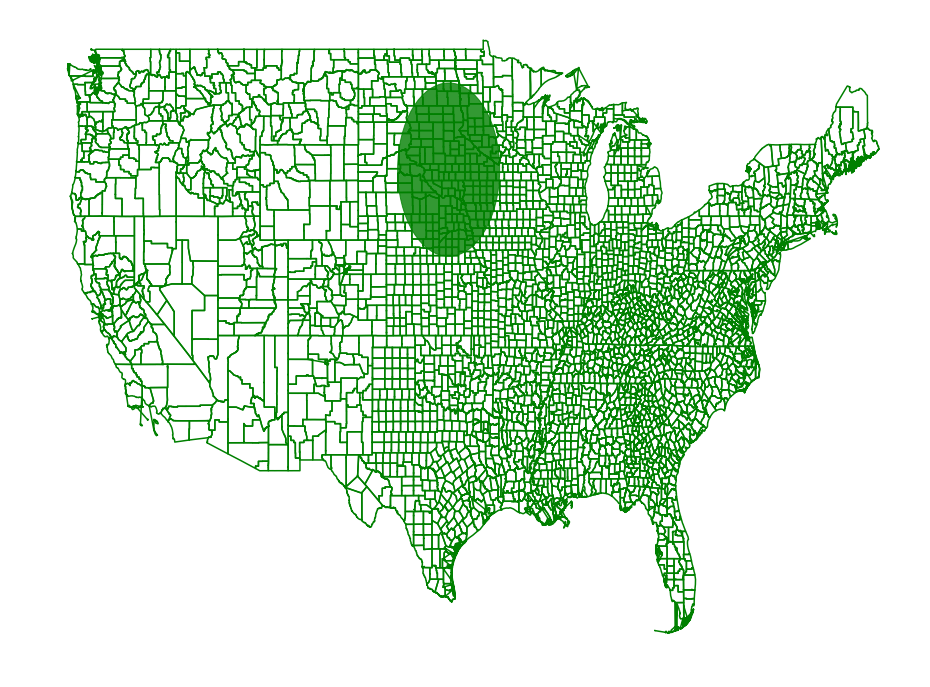
Region Scanning Part 2
Before we subsampled a set of regions and then scanned the sampled data
set to find a region of interest. In some cases we might want to scan a
set of regions, but with the constraint that some amount of overlap on a
range causes the region to be considered inside of that range. We can
encode these constraints into the range scanning, but we have to reduce
to a point set using a different procedure then before.
Now we use the region simplification code from the region simplification
page. We will scan all disks with radii between 24 miles and 240 miles
in diameter with an accuracy of 10 miles.
This function scans all the regions defined by the first argument and
measures the density of the regions set with the weights “weights2010”
and the regions set with the weights “weights2017”. Internally it
constructs approximations for the regions that alpha spatial error for
disks of size r_min to 2 * r_min. It then scans these approximations
and repeats with successively larger resolutions.
 pyscan
1.0
pyscan
1.0